👽 Cell phenotyping using a Hierarchical Prior Knowledge Table
1 2 3 4 | |
Running SCIMAP 1.3.14
1 2 | |
This method requires more work but is significantly more sensitive and scalable compared to clustering-based approaches.
To run the method, you'll need two main components:
To successfully execute the method, you will need:
- A
.csvfile containing manual gates. Should manual gates be absent, the algorithm will attempt to adjust the data by fitting two Gaussian distributions. Nonetheless, employing manual gating is recommended due to its heightened sensitivity. - A
.csvfile outlining a gating workflow strategy.
The execution of the algorithm is structured into three primary steps:
- Gate Identification: Utilize
sm.pl.gate_finderto identify the gates. - Data Rescaling: Apply
sm.pp.rescaleto adjust the data based on the identified gates. - Phenotyping: Process the rescaled data using
sm.tl.phenotypeto run the phenotyping algorithm.
Step 1: Define manual gates
1 | |
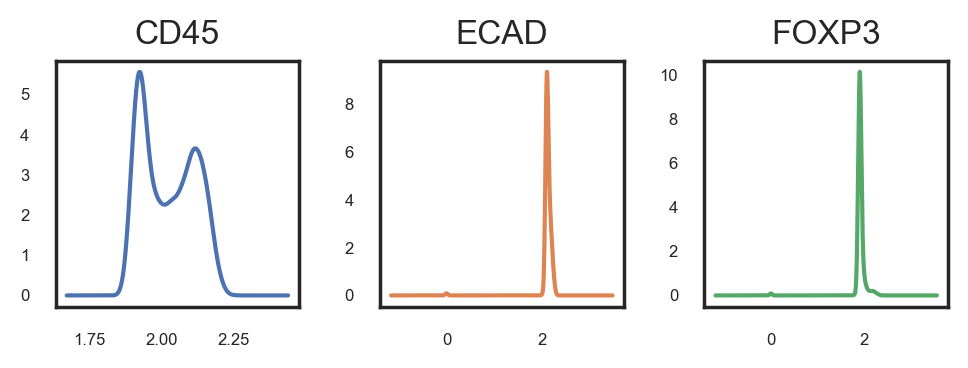
This method will launch a napari window displaying several layers, each representing a different gate. You can toggle these layers on and off with the goal of identifying the gate that most accurately captures the positive cells. It's crucial to zoom and pan through the image to ensure the selected gate encompasses all positive cells throughout the image. Bear in mind that achieving 100% accuracy might not be feasible, so a certain margin of error may need to be accepted. This process is iterative, meaning adjustments may be necessary even after initial phenotyping and analysis. Manual gating must be conducted independently for each marker and each image.
1 2 | |
1 | |
1 2 | |
| marker | exemplar-001--unmicst_cell | |
|---|---|---|
| 0 | ELANE | 7.80 |
| 1 | CD57 | 8.90 |
| 2 | CD45 | 6.40 |
| 3 | CD11B | 7.60 |
| 4 | SMA | 7.50 |
| 5 | CD16 | 6.50 |
| 6 | ECAD | 7.35 |
| 7 | FOXP3 | 7.40 |
| 8 | NCAM | 7.00 |
You'll observe that the first column lists the markers present in the dataset, while the second column specifies the gate, named after the specific image's ID found in adata.obs['imageid']. This is especially useful when dealing with datasets containing multiple images, as it allows for a distinct column for each image.
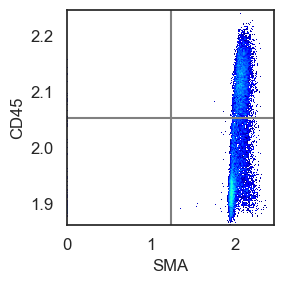
Although visual gating has proven to be the most sensitive method for us, you can also apply single and bi-marker gating approaches, similar to FACS, to assist in determining a threshold.
1 2 | |
We also use bimarker gating to identify a gate
1 | |
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/mpl_scatter_density/generic_density_artist.py:77: RuntimeWarning:
All-NaN slice encountered
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/mpl_scatter_density/generic_density_artist.py:82: RuntimeWarning:
All-NaN slice encountered
Step 2: Rescale the data
Here, we input a manual_gates.csv file into the gate parameter. This file contains gates determined visually with the help of the sm.pl.gate_finder function. For the markers specified in the manual_gates.csv file, the function will adjust the data so that cells exhibiting expression levels above the gate threshold are classified as positive for that marker, while those with expression levels below the threshold are deemed negative.
For markers not listed in the manual_gates.csv file, the function will employ an automatic approach to identify suitable gates by applying a Gaussian mixture model algorithm to the data.
1 2 3 | |
Scaling Image exemplar-001--unmicst_cell
Scaling ELANE
Scaling CD57
Scaling CD45
Scaling CD11B
Scaling SMA
Scaling CD16
Scaling ECAD
Scaling FOXP3
Scaling NCAM
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/scimap/preprocessing/rescale.py:105: FutureWarning:
Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
Step 3: Run the phenotyping algorithm
1 2 3 4 | |
| Unnamed: 0 | Unnamed: 1 | ELANE | CD57 | CD45 | CD11B | SMA | CD16 | ECAD | FOXP3 | NCAM | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | all | ECAD+ | pos | ||||||||
| 1 | all | Immune | pos | ||||||||
| 2 | all | SMA+ | pos | ||||||||
| 3 | Immune | NK cells | allpos | neg | allpos | ||||||
| 4 | Immune | Other myeloid cells | pos | ||||||||
| 5 | Immune | Treg | pos | ||||||||
| 6 | Other myeloid cells | Dendritic cells | allneg | allneg |
As it can be seen from the table above,
- The
first columnhas to contain the cell that are to be classified. - The
second columnindicates the phenotype a particular cell will be assigned if it satifies the conditions in the row. Column threeand onward represent protein markers. If the protein marker is known to be expressed for that cell type, then it is denoted by eitherpos,allpos. If the protein marker is known to not express for a cell type it can be denoted byneg,allneg. If the protein marker is irrelevant or uncertain to express for a cell type, then it is left empty.anyposandanynegare options for using a set of markers and if any of the marker is positive or negative, the cell type is denoted accordingly.
To give users maximum flexibility in identifying desired cell types, we have implemented various classification arguments as described above for strategical classification. They include
- allpos
- allneg
- anypos
- anyneg
- pos
- neg
pos : "Pos" looks for cells positive for a given marker. If multiple markers are annotated as pos, all must be positive to denote the cell type. For example, a Regulatory T cell can be defined as CD3+CD4+FOXP3+ by passing pos to each marker. If one or more markers don't meet the criteria (e.g. CD4-), the program will classify it as Likely-Regulatory-T cell, pending user confirmation. This is useful in cases of technical artifacts or when cell types (such as cancer cells) are defined by marker loss (e.g. T-cell Lymphomas).
neg : Same as pos but looks for negativity of the defined markers.
allpos : "Allpos" requires all defined markers to be positive. Unlike pos, it doesn't classify cells as Likely-cellType, but strictly annotates cells positive for all defined markers.
allneg : Same as allpos but looks for negativity of the defined markers.
anypos : "Anypos" requires only one of the defined markers to be positive. For example, to define macrophages, a cell could be designated as such if any of CD68, CD163, or CD206 is positive.
anyneg : Same as anyneg but looks for negativity of the defined markers.
1 | |
Phenotyping ECAD+
Phenotyping Immune
Phenotyping SMA+
-- Subsetting Immune
Phenotyping NK cells
Phenotyping Other myeloid cells
Phenotyping Treg
-- Subsetting Other myeloid cells
Phenotyping Dendritic cells
Consolidating the phenotypes across all groups
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/scimap/tools/phenotype_cells.py:174: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/scimap/tools/phenotype_cells.py:290: FutureWarning:
DataFrame.fillna with 'method' is deprecated and will raise in a future version. Use obj.ffill() or obj.bfill() instead.
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/scimap/tools/phenotype_cells.py:290: FutureWarning:
Downcasting object dtype arrays on .fillna, .ffill, .bfill is deprecated and will change in a future version. Call result.infer_objects(copy=False) instead. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
1 2 | |
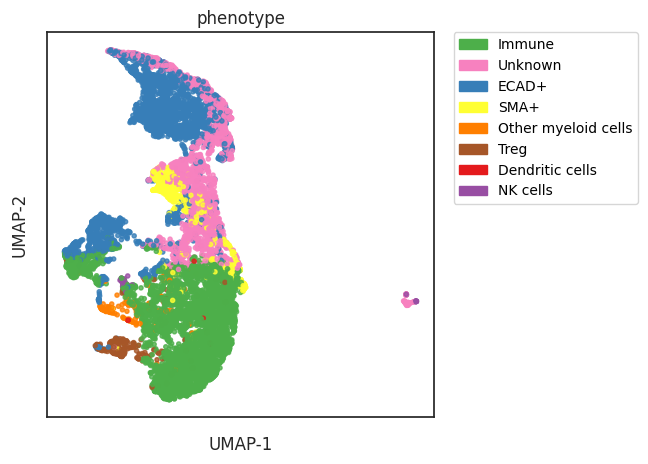
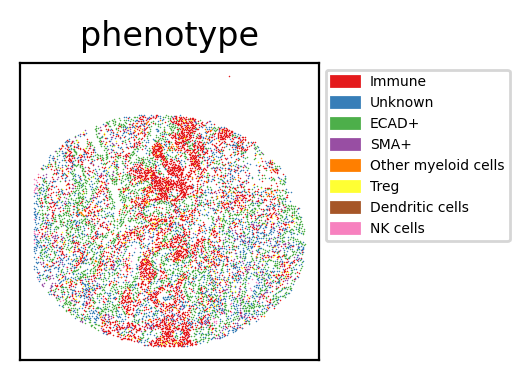
phenotype
Immune 4746
ECAD+ 3015
Unknown 2278
SMA+ 602
Treg 282
Other myeloid cells 193
NK cells 66
Dendritic cells 19
Name: count, dtype: int64
1 | |
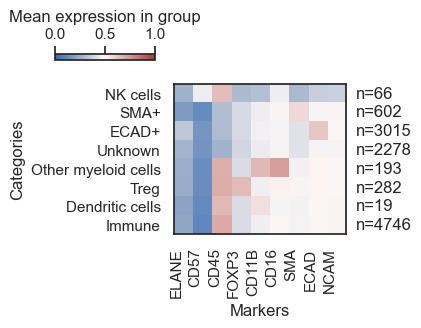
Visualisation of the Results
1 2 | |
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/scimap/plotting/heatmap.py:312: UserWarning:
This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
1 2 | |
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning:
n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
1 | |
/Users/aj/miniconda3/envs/scimap/lib/python3.10/site-packages/scimap/plotting/umap.py:267: UserWarning:
No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
1 2 | |
1 2 3 4 5 6 7 8 9 10 11 12 13 | |
Save Results
1 2 | |
1 | |