Home
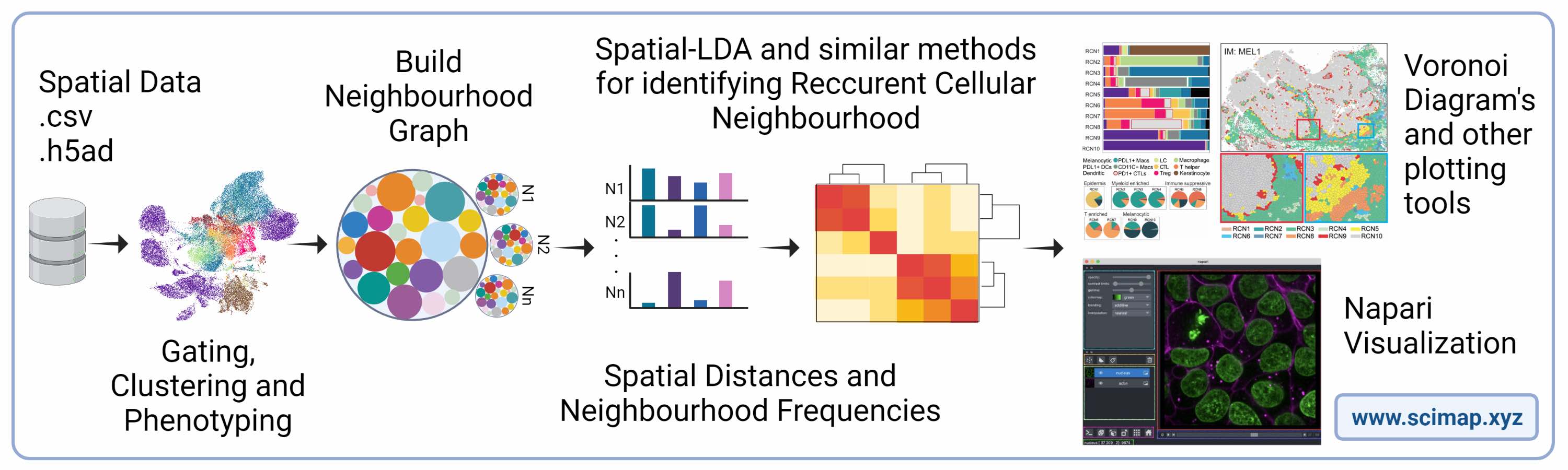
Scimap is a scalable toolkit for analyzing spatial molecular data. The underlying framework is generalizable to spatial datasets mapped to XY coordinates. The package uses the anndata framework making it easy to integrate with other popular single-cell analysis toolkits. It includes preprocessing, phenotyping, visualization, clustering, spatial analysis and differential spatial testing. The Python-based implementation efficiently deals with large datasets of millions of cells.
Scimap operates on segmented single-cell data derived from imaging data using tools such as cellpose or MCMICRO. The essential inputs for SCIMAP are: (a) a single-cell expression matrix and (b) the X and Y coordinates for each cell. Additionally, multi-stack OME-TIFF or TIFF images can be optionally provided to enable visualization of the data analysis on the original raw images.
Scimap development was led by Ajit Johnson Nirmal, Harvard Medical School.
Check out other tools from the Nirmal Lab.
Citing scimap
Nirmal et al., (2024). SCIMAP: A Python Toolkit for Integrated Spatial Analysis of Multiplexed Imaging Data. Journal of Open Source Software, 9(97), 6604, https://doi.org/10.21105/joss.06604
Funding
This work was supported by the following NIH grant K99-CA256497